Cookiecutter Docker Science
Data scientists do many machine learning or data mining tasks. For such data engineering tasks, researchers apply various tools and system libraries, which are constantly updated. Unfortunately installing and updating them cause problems in local environments. Even when we work in hosting environments such as EC2, we are not free from this problem. Some experiments succeeded in an instance but failed in another one, since library versions of each EC2 instances could be different.
By contrast, we can creates the identical environments with Docker in which needed tools with the correct versions are installed without changing system libraries in host machines.
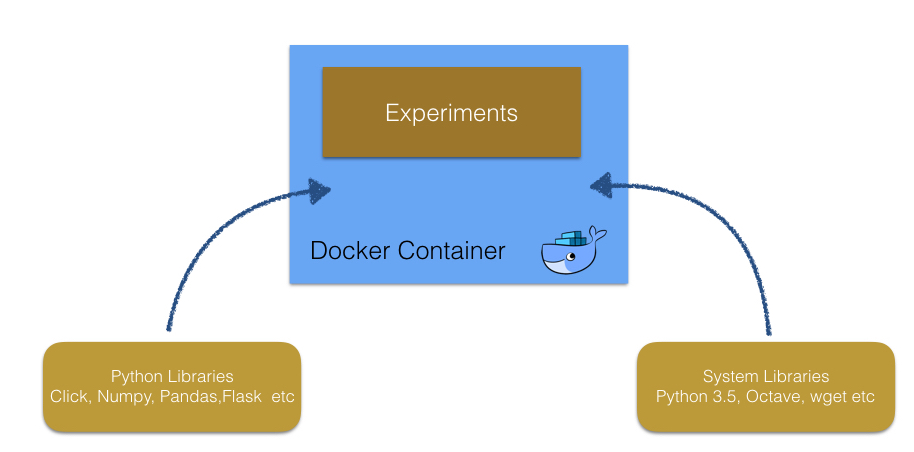
This aspect of Docker is important for reproducibility of experiments, and keep the projects in continuous integration systems.
However, working in a Docker containers is troublesome. Adding a new library into requirements.txt
or Dockerfile does not installed as if a local machine. Specifically we need to create a Docker image and the container each time.
We also need to add port forward settings to see server responses such as Jupyter Notebook UI launch in Docker container from our local environments.
Cookiecutter Docker Science provides utilities to make working in Docker container simple.
This project is a tiny template for machine learning projects developed in Docker environments. In machine learning tasks, projects glow uniquely to fit target tasks, but in the initial state, most directory structure and targets in Makefile are common. Cookiecutter Docker Science generates initial directories which fits simple machine learning tasks.
Cycle of a project
Generally machine learning projects consist of three phases (Experiments, Code simplification, and Deployment). The following is the image of the cycle of a machine learning project.
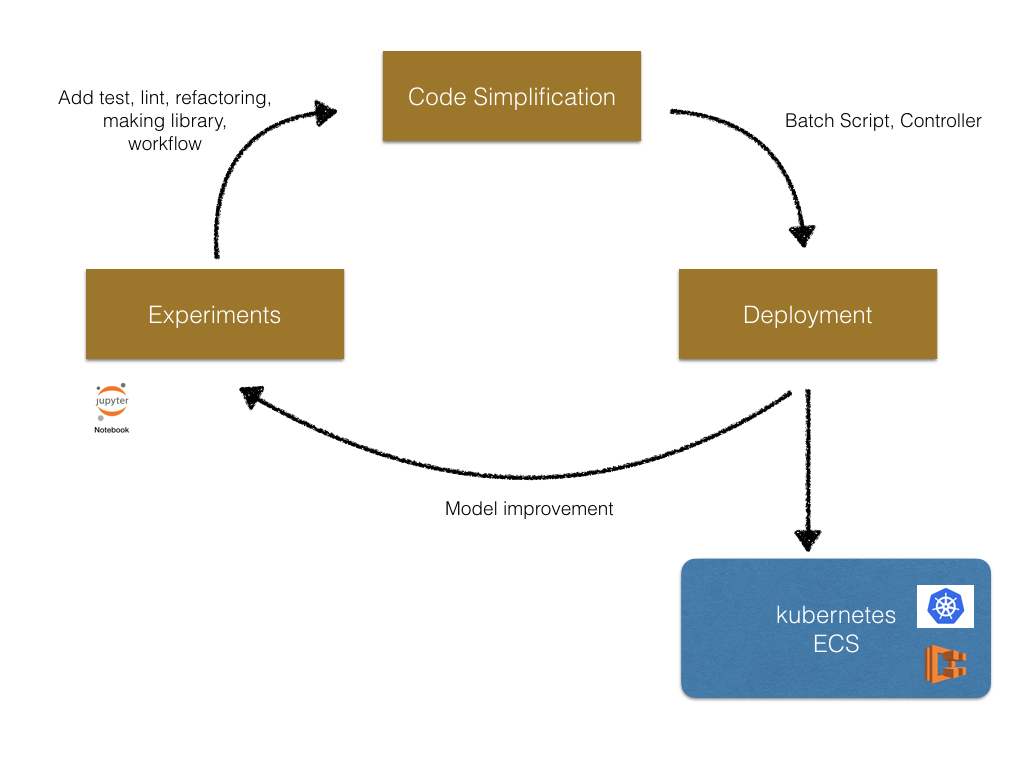
In the begging we do experiments in Jupyter Notebook interactively. Then we simplify the code written in the notebooks. In this step, add test, refactor code, apply linter, make library and CI. After we finished the code simplification, we deploy the model to production use. In this phase, we add batch scripts or service using the library created at the previous step. After deployment, we continue the improvement of model in Jupyter Notebook again.
Machine learning project with Docker containers
Ideally, we should work in a Docker container in every step of the machine learning projects generated by the same Dockerfile, since we can start code simplification and deployment seamlessly. The following is the image.
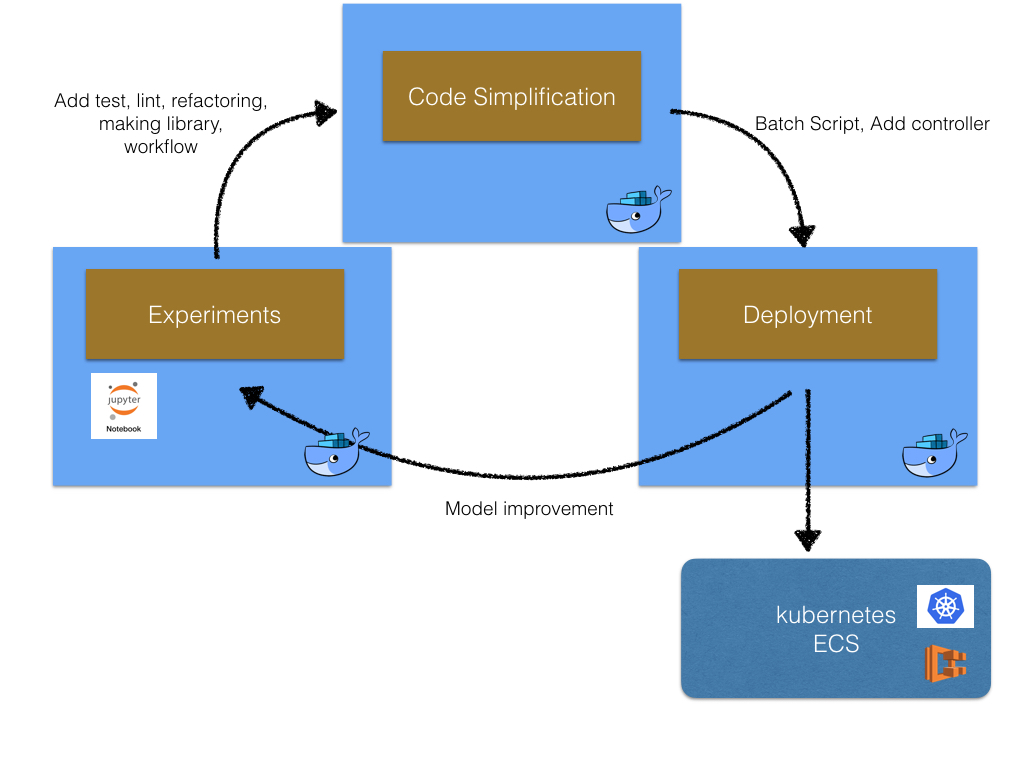
Cookiecutter Docker Science supports the machine learning projects from the experiments to deployment with Docker container.
Usage
This section describes the usage of Cookiecutter Docker Science.
Requirements
Cookiecutter Docker Science need the followings.
- Python 2.7 or Python 3.5
- Cookiecutter 1.6 or later
- Docker version 17 or later
Generate new project
To generate project with the Cookiecutter Doccker Science template, please run the following command.
$ cookiecutter git@github.com:docker-science/cookiecutter-docker-science.git
Then the cookiecutter asks for several questions on generated project as follows.
$cookiecutter git@github.com:docker-science/cookiecutter-docker-science.git
project_name [project_name]: food-image-classification
project_slug [food_image_classification]:
jupyter_host_port [8888]:
description [Please Input a short description]: Classify food images into several categories
data_source [Please Input data source in S3]: s3://research-data/food-images
Create Docker Container
To create a Docker image and container and then launch Jupyter Notebook in the container,
we run make init-docker and make create-container commands in the generated project directory.
$ cd food-image-classification
$ make init-docker
$ make create-container
Now we are in Docker container.
Run Jupyter Notebook
To launch Jupyter Notebook, we run the following command.
$ make jupyter
Directory structure
When we generate a project with Cookiecutter Docker Science, the project has the following files and directories.
├── Makefile <- Makefile contains many targets such as create docker container or
│ get input files.
├── config <- This directory contains configuration files used in scripts
│ │ or Jupyter Notebook.
│ └── jupyter_config.py
├── data <- data directory contains the input resources.
├── docker <- docker directory contains Dockerfile.
│ ├── Dockerfile <- Base Dockerfile contains the basic settings.
│ ├── Dockerfile.dev <- Dockerfile for experiments this Docker image is derived from the base Docker image.
│ │ This Docker image does not copy the files and directory but used mount the top
│ │ directory of the host environments.
│ └── Dockerfile.release <- Dockerfile for production this Docker image is derived from the base Docker image.
│ The Docker image copy the files and directory under the project top directory.
├── model <- model directory store the model files created in the experiments.
├── my_data_science_project <- cookie-cutter-docker-science creates the directory whose name is same
│ │ as project name. In this directory users puts python files used in scripts
│ │ or Jupyter Notebook.
│ └── __init__.py
├── notebook <- This directory sotres the ipynb files saved in Jupyter Notebook.
├── requirements.txt <- Libraries needed to run experiments. The library listed in this file
│ are installed in the Docker container.
├── scripts <- Users add the script files to generate model files or run evaluation.
└── tests <- tests directory stores test codes and the fixture files.
Edit codes with preferred editors
The top level files and directories are mounted into the generated Docker container, and therefore when the files are edited in local machines, the changes are immediately reflected in the Docker container. All we need in Docker containers is just run the scripts.
As the result, we can edit files any editors as we like. Some data analyst edits with PyCharm other software engineers simplify the results of Jupyter Notebook with vim as the following image.
Consistent support
As described in the previous sections, machine learning projects consist of three phases (experiments, code simplification, and deployment). Cookiecutter Docker Science supports machine learning projects in all three phases.
Experiments
Projects start from exploratory experiments. This phase data scientists analysis given data in Jupyter Notebook interactively.
Cookiecutter Docker Science launches the Jupyter Notebook server in the container with make jupyter command. Note that
port forward setting to connect the port Docker container to host PC is setup by Cookiecutter Docker Science.
Users create notebooks in notebook directory and store the results of analysis.
Code simplification
In the code simplification phase, users extract components used in the experiments to the library directory. The library codes are stored in the library directory (commonly project name directory) generated by Cookiecutter Docker Science.
Then software engineers refactor codes and add tests. The test files are stored in tests directory.
The flows to generate the model files are added in Makefile as the targets.
Adding the make targets we can generate the model with just make command.
Deployment
After code simplification phase, we deploy the model to the service or batch script.
We can make use of the project since the directories in the projects
are the same and we are able to generate model with the make command.
Specifically we can create the production Docker images reducing libraries or settings for development uses,
just running make init-docker specifying a environment variable MODE to release as make init-docker MODE=release.
Projects apply Cookiecutter Docker Science
- Udacity Deep Reinforcement Learning Nanodegree Project: Reacher by CCThompson82
- sobel filter on images by genkioffice
License
Apache version 2.0
